Research
Prediction of the three-dimensional structure and function of proteins from pathogenic bacteria
Ab initio molecular modeling techniques have been used for in silico structural genomics initiatives for the prediction of the structure and function of proteins from pathogenic bacteria. This initiative has led to obtain structural and functional information on about 70 Streptococcus mutans proteins (Bizai et al., manuscript in preparation) and 90 Pseudomonas aeruginosa proteins (Caprari et al., manuscript in preparation), some of which could be used as molecular targets for the development of new antibacterial drugs.
Development of tools for ligand binding site prediction and drug design
Structure-function relationship in Huntingtin
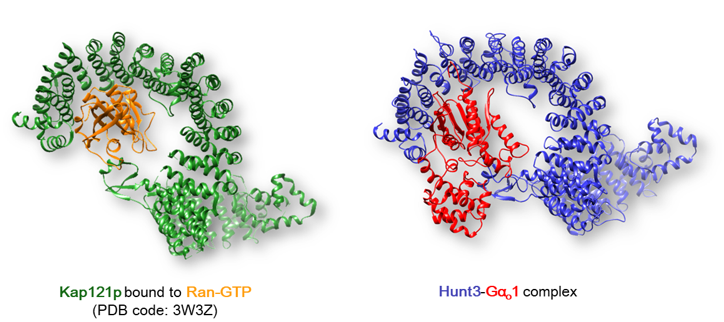
Huntington's disease (HD) is a rare neurodegenerative disorder elicited by an increase of CAG repeats within the coding region of a protein called huntingtin (htt). Its high molecular weight together with the presence of a disordered regions hinders crystallization and X-ray diffraction studies. Through the use of several methods htt regions predicted to fold into ordered domains have been detected, providing a putative function for each domain (Brandi et al., 2017). The third ordered domain (Hunt3) has shown structural similarity with a Karyopherin, a protein involved in nuclear import by interacting with GTP-binding proteins. Surprisingly, the analysis of the htt interactome has revealed that it interacts with several GTP binding proteins, such as Gαo1. Molecular docking simulations have confirmed that Hunt3 can be considered the putative interaction site between htt and G proteins (Brandi et al., 2017).
Structure-function computational analysis of membrane transporters involved in human diseases
The solute carrier (SLC) transporter superfamily is particularly relevant in human physiology, with important targets identified in neuroscience, metabolism, inflammatory diseases and oncology (César-Razquin et al., 2015; Lin et al., 2015). Protein modeling, molecular docking and molecular dynamics simulations are used to understand the structure-function relationship of these transporters. In particular, iron transporters and mitocondrial carriers are being studied (Tortosa et al., 2017).

Development of software tools for the prediction of protein function
An ongoing activity is the development of software tools for local structural comparison of protein structures, for the prediction of the catalytic activity of proteins of unknown function and for the identification of ligand binding sites in proteins. ASSIST (which stands for Active Site Similarity Search Tool) is a software for the prediction of the catalytic activity of proteins. The ASSIST algorithm is based on the geometric hashing technique and allows to compare an input protein structure with a database of about 1000 catalytic sites with execution times of the order of tens of seconds. The program has a "user friendly" graphic interface for the input procedure and for the analysis of the output. Tests carried out on about 60 randomly selected protein structures indicated that ASSIST is able to correctly predict the catalytic activity of a protein in almost all of the cases analyzed (Caprari et al., 2013). ASSIST has been developed in Java and thus can be run on any hardware platform. ASSIST can be downloaded on the Software section.
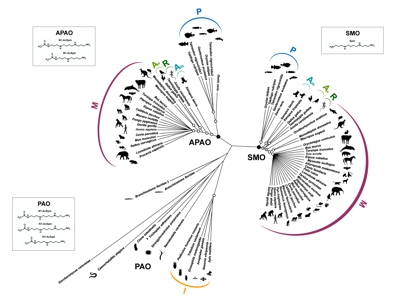
Structural basis of the catalytic mechanism and substrate specificity of polyamine oxidases
The catalytic mechanism and substrate specificity of the vertebrate polyamine oxidases spermine oxidase (SMO) and acetilpoliammino oxidase (APAO) are studied using molecular modeling techniques, rational design of site-specific mutants and molecular docking simulations combined with experimental studies. These studies have allowed us to identify residues involved in the binding of the substrates to SMO and APAO (Tavladoraki et al., 2011; Cervelli et al., 2012) and to clarify the molecular bases of the different substrate specificity of these two enzymes (Polticelli et al., 2012; Cervelli et al., 2013). It has been also possible to understand the role of specific residues in the catalytic mechanism of this family of enzymes, through the design of site-specific mutants and the structural characterization of these mutants by X-ray diffraction (Tavladoraki et al., 2011; Fiorillo et al., 2011).
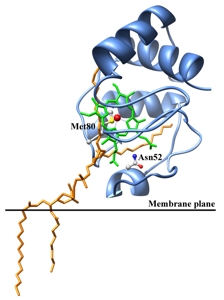
Molecular basis of the involvement of cytochrome c in the apoptotic process
Modeling studies and molecular docking on the complex Cyt c- cardiolipin allowed to hypothesize the possible binding mode of the lipid to Cyt c. The formation of this complex leads to the conversion of Cyt c from a protein active in electron transfer to an enzyme with peroxidase activity involved in the early events of the apoptotic process (Ascenzi et al., 2011; Patriarca et al., 2012). Furthermore the role of effectors, such as ATP and sodium chloride, and of the basic residues of the protein in the modulation of the interaction Cyt c-cardiolipin has been investigated (Sinibaldi et al., 2011; Patriarca et al., 2013).